Welcome to mOWL’s documentation!
mOWL is a Python library for Machine Learning with Ontologies. Here you can find functionalities to manipulate ontologies and use them as data for several methods that generate embeddings of ontology entities.
Background
Several methods to generate vector representations have been developed and identified in: Semantic similarity and machine learning with ontologies. In this work, several tutorials and slides have been created for users that are interested on ontologies and machine learning methods. The tutorials can be found as Jupyter notebooks at this link to the notebooks. The slides can be found at this link to the slides.
mOWL: Python library for machine learning with biomedical ontologies was developed to provide a standardized framework to use the already existing methods and to ease the development of new ones.
Some of those tutorials can be found in mOWL in the Paper study with mOWL section and, eventually, more examples will be available directly in mOWL.
Getting started
mOWL can be installed from source code or from PyPi. For more details on installation check out the how to Installation and Setup section of the project.
Note
This version of documentation corresponds to mOWL-0.3.0.
mOWL, JPype and the JVM
mOWL is a Python library. However, it interfaces the OWLAPI, which is written in Java. For that reason, mOWL uses JPype to use the JVM and access Java code from Python scripts.
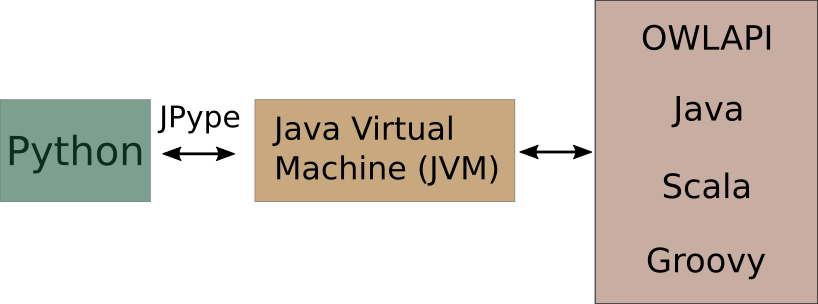
In order to use mOWL with all its functionalities, the Java Virtual Machine must be started. We can do that in the following way:
import mowl
mowl.init_jvm("2g")
In the above piece of code, we specify the amount of memory given to the JVM. The memory parameter (2g in the example) corresponds to the parameter “-Xmx” for the JVM initialization step. For more information about the JVM memory management please follow this link.
Note
The function init_jvm can only be called once during running time. This means that the JVM cannot be restarted and this is a limitation of JPype as stated in this section of their documentation.
License
The package is released under the BSD 3-Clause License.
Citation
If you used mOWL in your work, please consider citing this article.
@article{10.1093/bioinformatics/btac811,
author = {Zhapa-Camacho, Fernando and Kulmanov, Maxat and Hoehndorf, Robert},
title = "{mOWL: Python library for machine learning with biomedical ontologies}",
journal = {Bioinformatics},
year = {2022},
month = {12},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btac811},
url = {https://doi.org/10.1093/bioinformatics/btac811},
note = {btac811},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btac811/48438324/btac811.pdf},
}